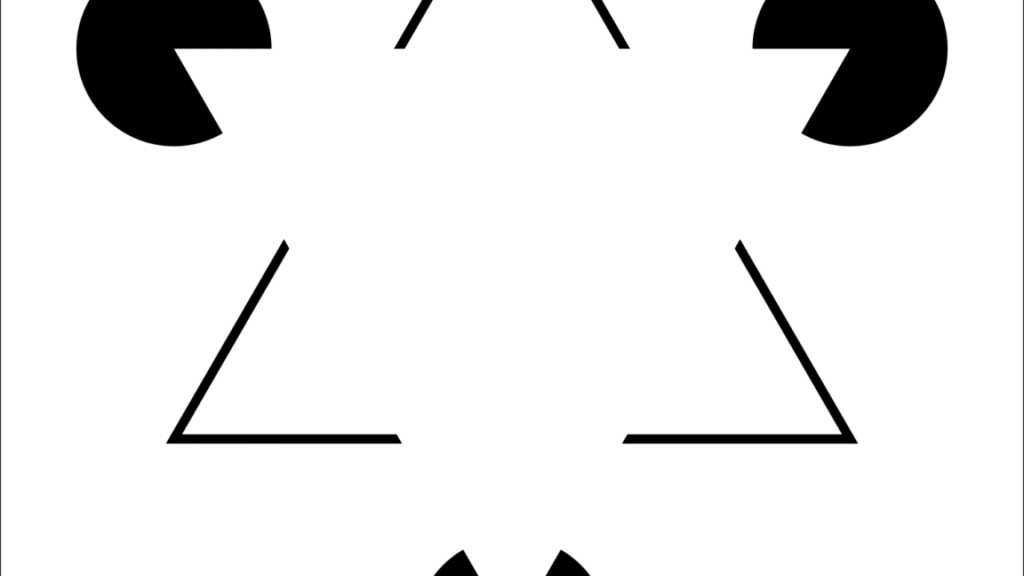
The neurons that let us see what isn’t there
Earlier work had hinted at such cells, but Shin and colleagues show systematically that they’re not rare oddballs—they’re a well-defined, functionally important subpopulation. “What we didn’t know is that these neurons drive local pattern completion within primary visual cortex,” says Shin. “We showed that those cells are causally involved in this pattern completion process that we speculate is likely involved in the perceptual process of illusory contours,” adds Adesnik.
Behavioral tests still to come
That doesn’t mean the mice “saw” the illusory contours when the neurons were artificially activated. “We didn’t actually measure behavior in this study,” says Adesnik. “It was about the neural representation.” All we can say at this point is that the IC-encoders could induce neural activity patterns that matched what imaging shows during normal perception of illusory contours.
“It’s possible that the mice weren’t seeing them,” admits Shin, “because the technique has involved a relatively small number of neurons, for technical limitations. But in the future, one could expand the number of neurons and also introduce behavioral tests.”
That’s the next frontier, Adesnik says: “What we would do is photo-stimulate these neurons and see if we can generate an animal’s behavioral response even without any stimulus on the screen.” Right now, optogenetics can only drive a small number of neurons, and IC-encoders are relatively rare and scattered. “For now, we have only stimulated a small number of these detectors, mainly because of technical limitations. IC-encoders are a rare population, probably distributed through the layers [of the visual system], but we could imagine an experiment where we recruit three, four, five, maybe even 10 times as many neurons,” he says. “In this case, I think we might be able to start getting behavioral responses. We’d definitely very much like to do this test.”
Nature Neuroscience, 2025. DOI: 10.1038/s41593-025-02055-5
Federica Sgorbissa is a science journalist; she writes about neuroscience and cognitive science for Italian and international outlets.
The neurons that let us see what isn’t there Read More »